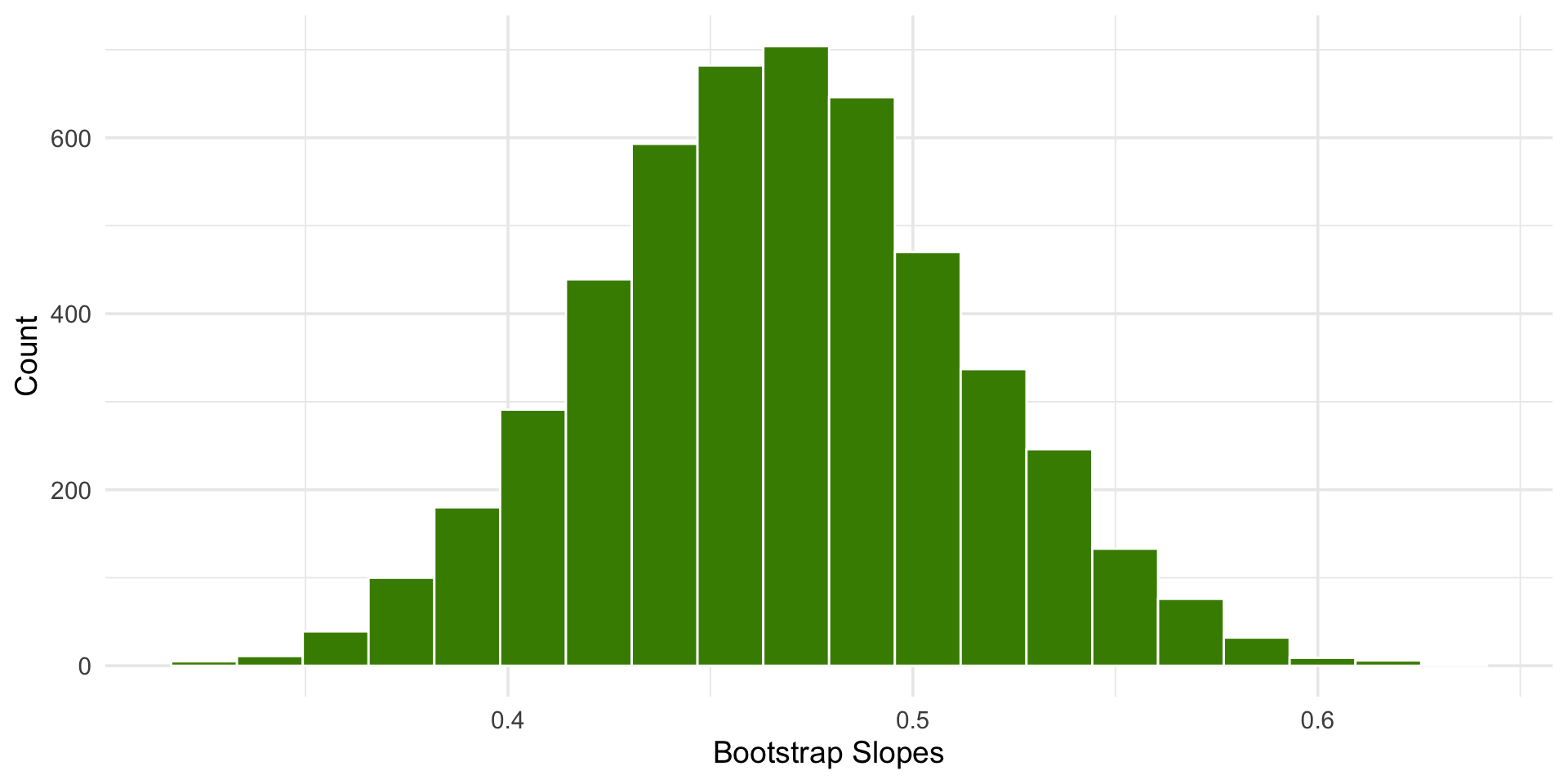
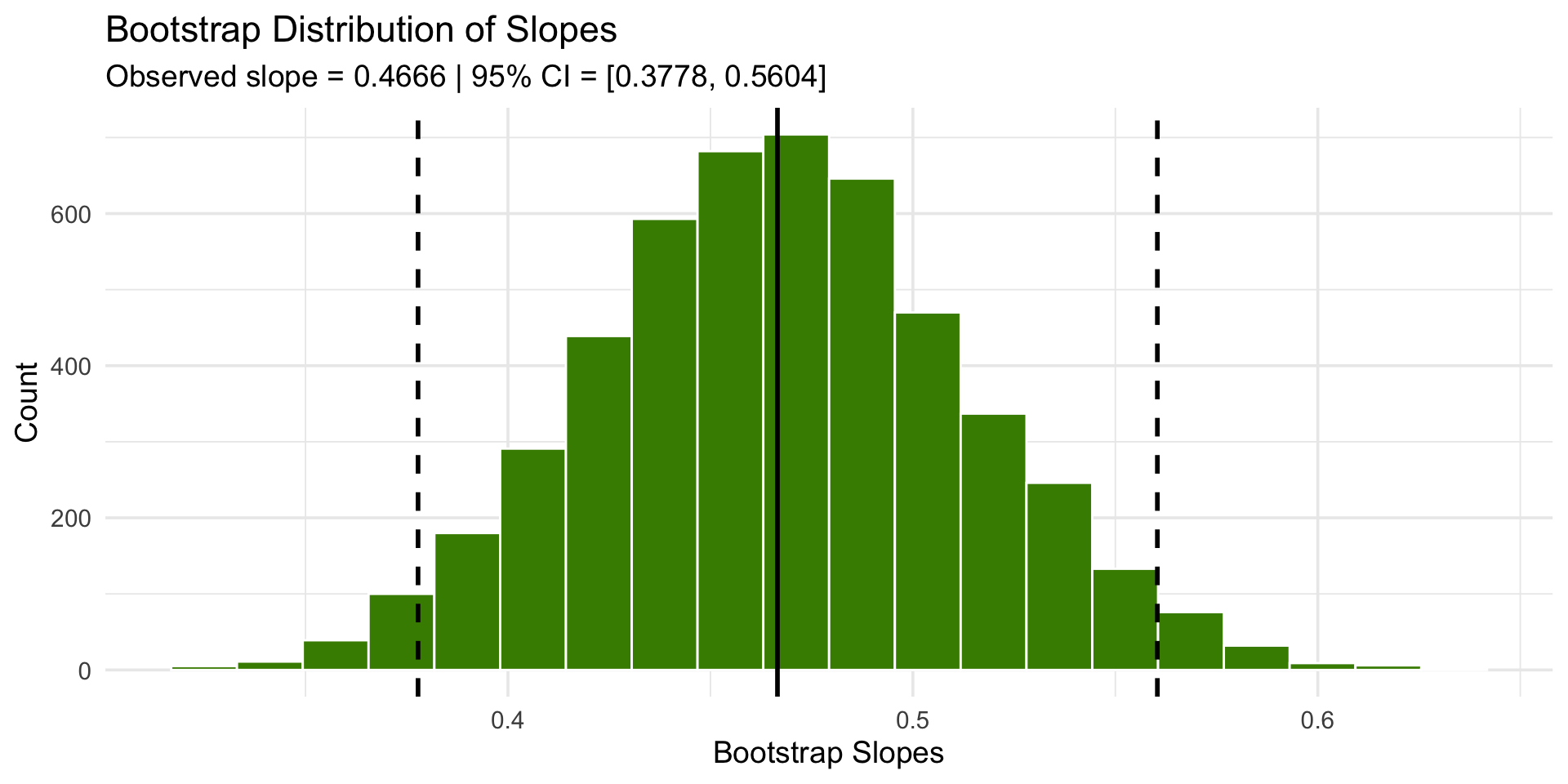
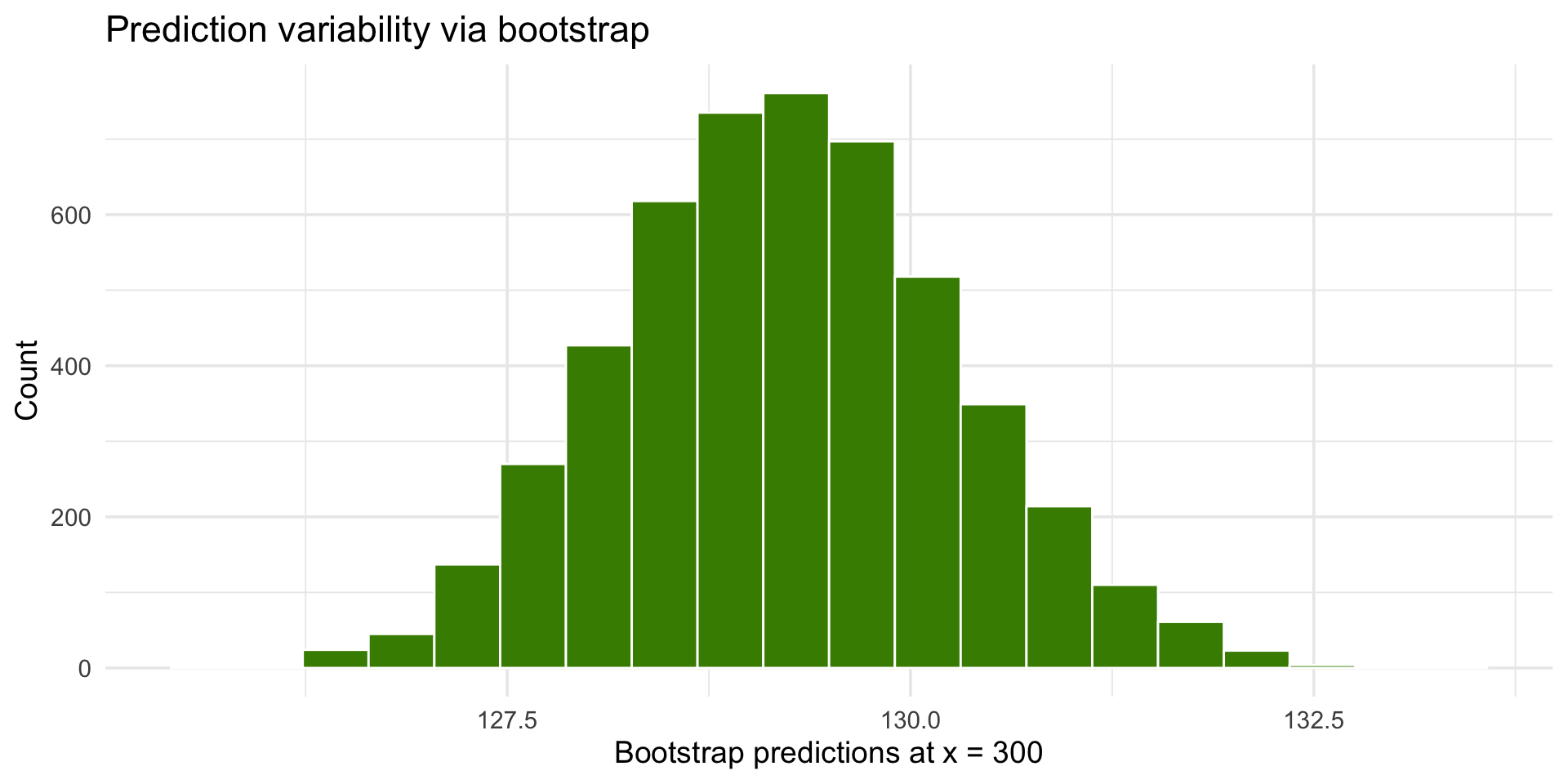
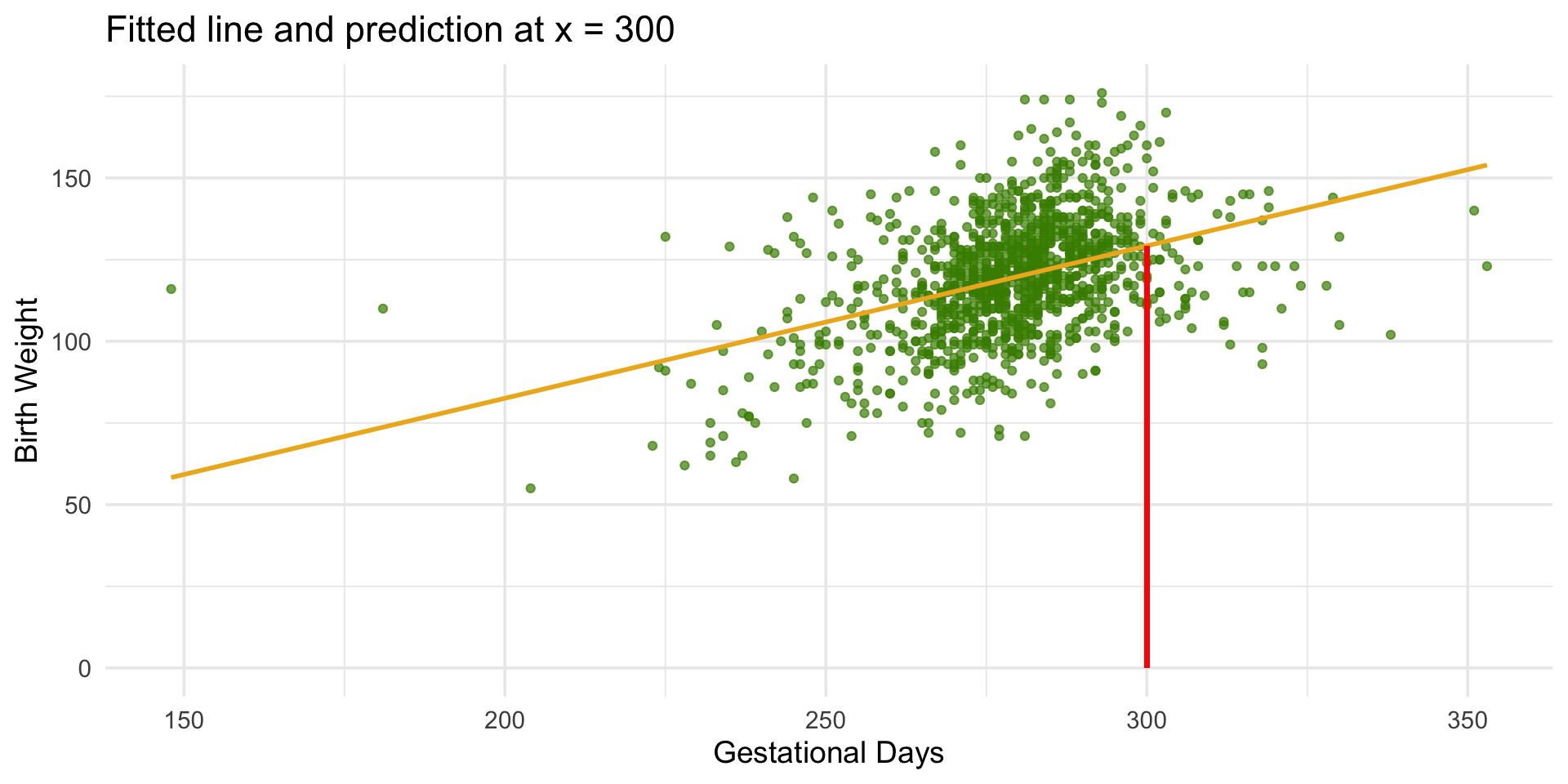
library(tidyverse)
library(infer)
library(broom)
baby <- read_csv("data/baby.csv", show_col_types = FALSE)
glimpse(baby)Rows: 1,174
Columns: 6
$ `Birth Weight` <dbl> 120, 113, 128, 108, 136, 138, 132, 120, 14…
$ `Gestational Days` <dbl> 284, 282, 279, 282, 286, 244, 245, 289, 29…
$ `Maternal Age` <dbl> 27, 33, 28, 23, 25, 33, 23, 25, 30, 27, 32…
$ `Maternal Height` <dbl> 62, 64, 64, 67, 62, 62, 65, 62, 66, 68, 64…
$ `Maternal Pregnancy Weight` <dbl> 100, 135, 115, 125, 93, 178, 140, 125, 136…
$ `Maternal Smoker` <lgl> FALSE, FALSE, TRUE, TRUE, FALSE, FALSE, FA…